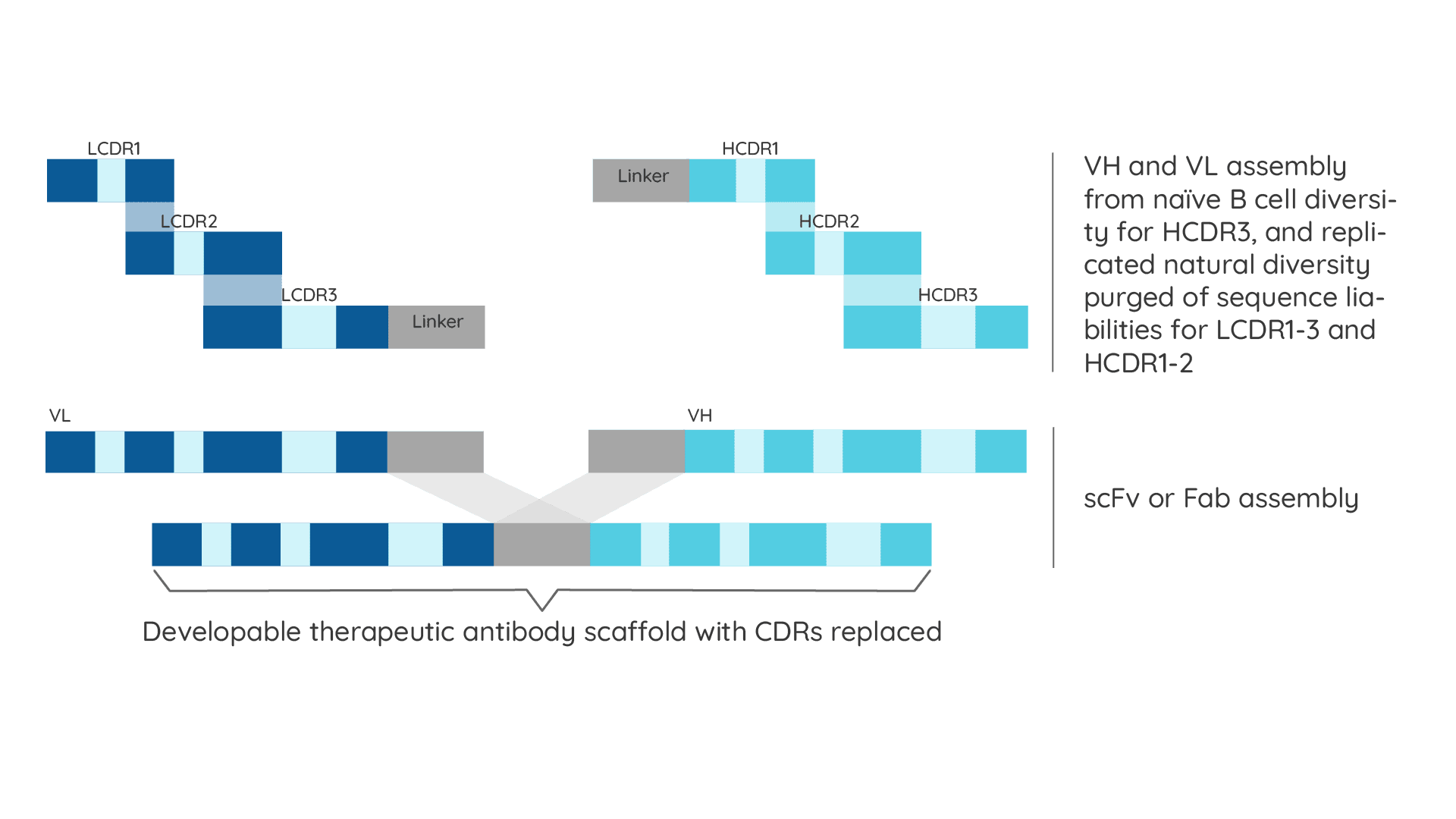
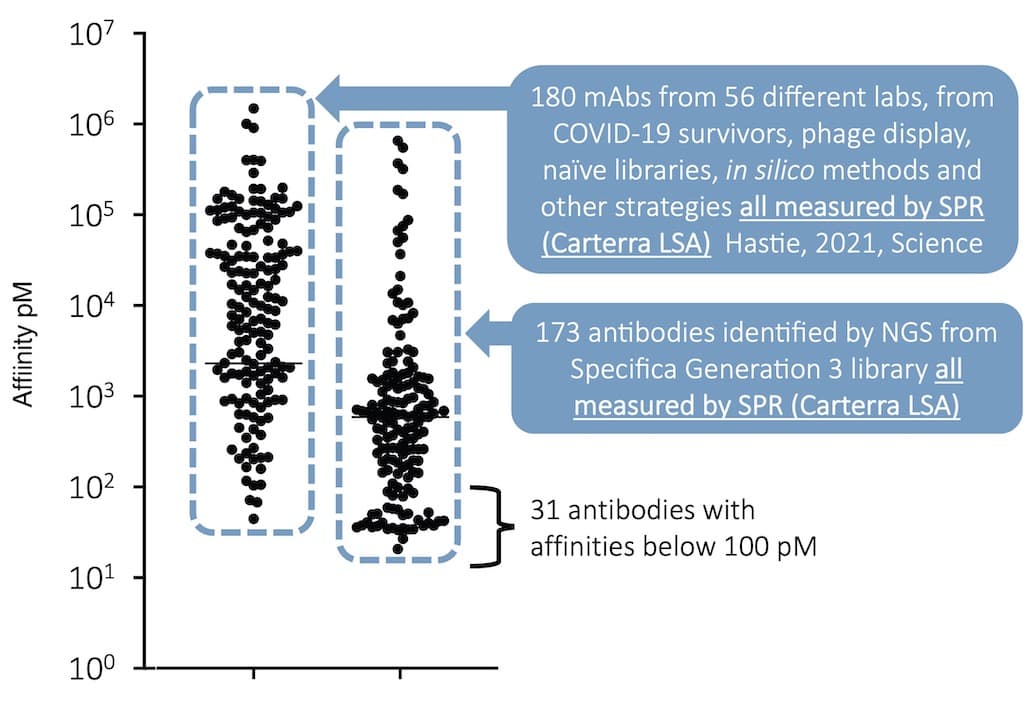
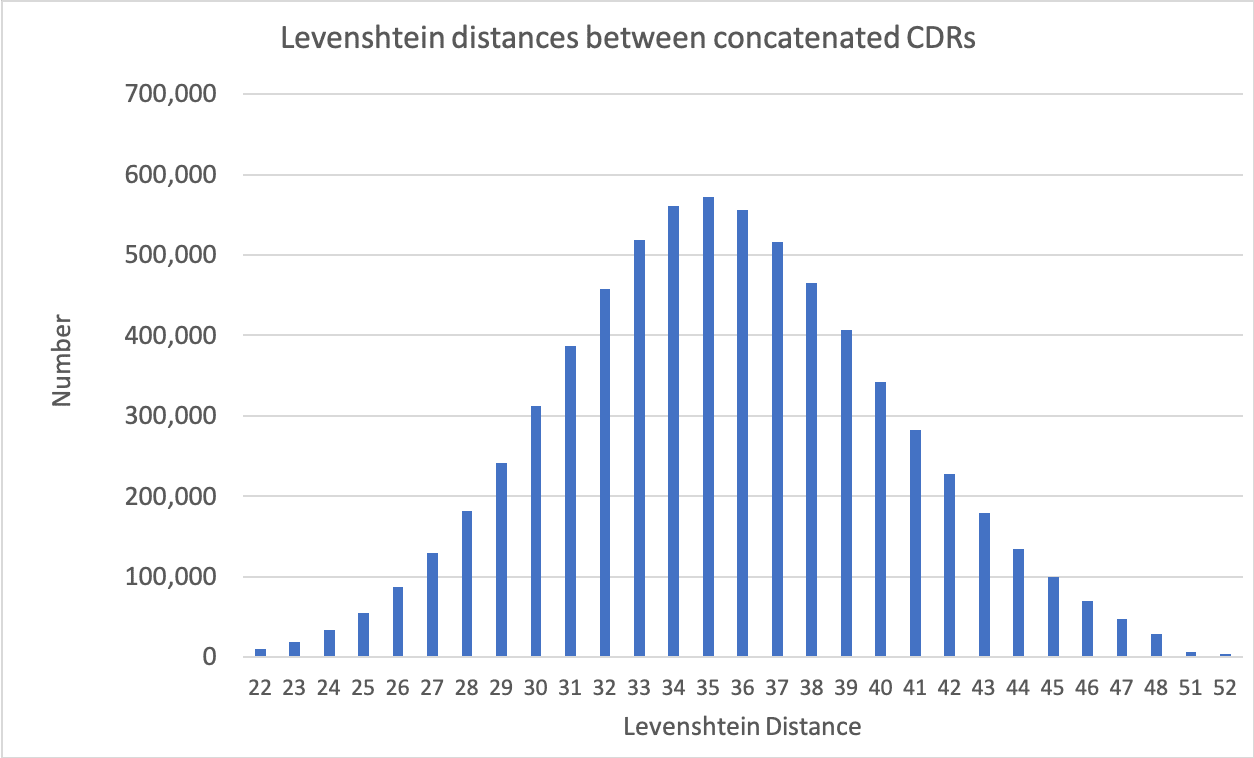
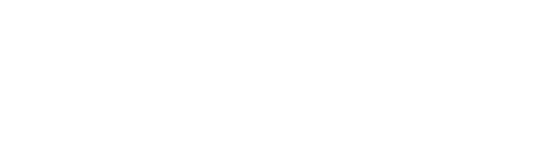Specifica’s Generation 3 Library Platform has quality built in by design. Developability, extremely high diversity, and the exclusion of sequence liabilities are intrinsic to the four sub-libraries making up the Platform, each of which is based on a therapeutic antibody scaffold chosen for its biophysical properties, lack of liabilities and germline gene variety.
Diversity is all derived from natural CDR sequences: HCDR3s are amplified directly from purified B cells, while replicated natural CDRs, derived from Specifica’s databases, are used for the rest. The same unique HCDR3 diversity can be repackaged into different formats, including VHH, scFv and Fab. In the case of scFv and Fab, libraries can also be made with common light chains as well as traditionally.