
Affinity maturation and antibody lead
Specifica, an IQVIA business, improves antibody leads using our proprietary Generation 3 technology.
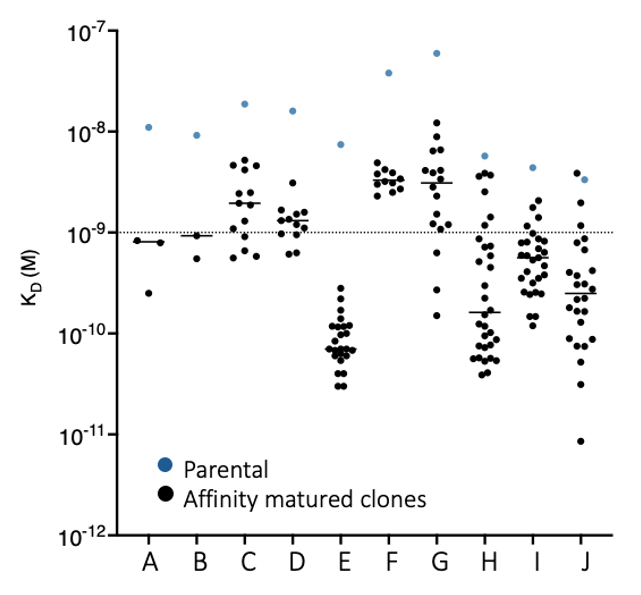
While antibodies from in vitro or in vivo methods may show desired specificity, they often lack optimal properties. Our most requested service —affinity maturation—routinely delivers 10- to 27,000-fold improvements over parental affinity.
We also optimize pH sensitivity, developability and species cross-reactivity through our platform and deep expertise.

| Format | Target CDRs | Mean improvement (fold) | Best improvement (fold) | |
| A | scFV-yeast | HCDR1-3 | 14 | 33 |
| B | scFV-yeast | LCDR1-3 & HCDR1-3 | 14 | 26 |
| C | scFV-yeast | LCDR1-3 & HCDR1-3 | 106 | 248 |
| D | scFV-yeast | LCDR1-3 & HCDR1-2 | 12 | 16 |
| E | scFV-yeast | LCDR1-3 & HCDR1-2 | 61 | 397 |
| F | scFV-yeast | LCDR1-3 & HCDR1-2 | 48 | 147 |
| G | scFV-yeast | LCDR1-2 & HCDR1-3 | 11 | 37 |
| H | scFV-yeast | LCDR1-2 & HCDR1-3 | 37 | 391 |
| I | scFV-yeast | LCDR1-2 & HCDR1-3 | 1076 | 27397 |
| J | scFV-yeast | LCDR1-3 & HCDR1-3 | 8 | 124 |
| K | scFV-yeast | LCDR1-3 & HCDR1-3 | 7 | 24 |
| L | scFV-yeast | LCDR1-3 & HCDR1-3 | 366 | 15000 |
| M | scFV-yeast | HCDR1-3 | 7 | 36 |
| M | scFV-yeast | HCDR1-3 | 1.4 | 10 |
Frequently asked questions:
Is the affinity improvement guaranteed?
Yes, Specifica guarantees at least a 10-fold improvement and routinely achieves more. If not achieved, the delivery fee is waived.
What makes Specifica's affinity maturation platform unique?
We leverage natural human diversity to enhance “humanness” and avoid introducing problematic CDR sequences. Our two-stage process explores vast diversity (≥10¹⁸) while preserving epitope recognition.
Can the maturation project be customized?
Absolutely. We tailor designs to your needs—whether limiting which CDRs are diversified or specifying mutations at particular amino acid positions.
Specifica applies the expertise of our Generation 3 library platform to improve your antibodies:
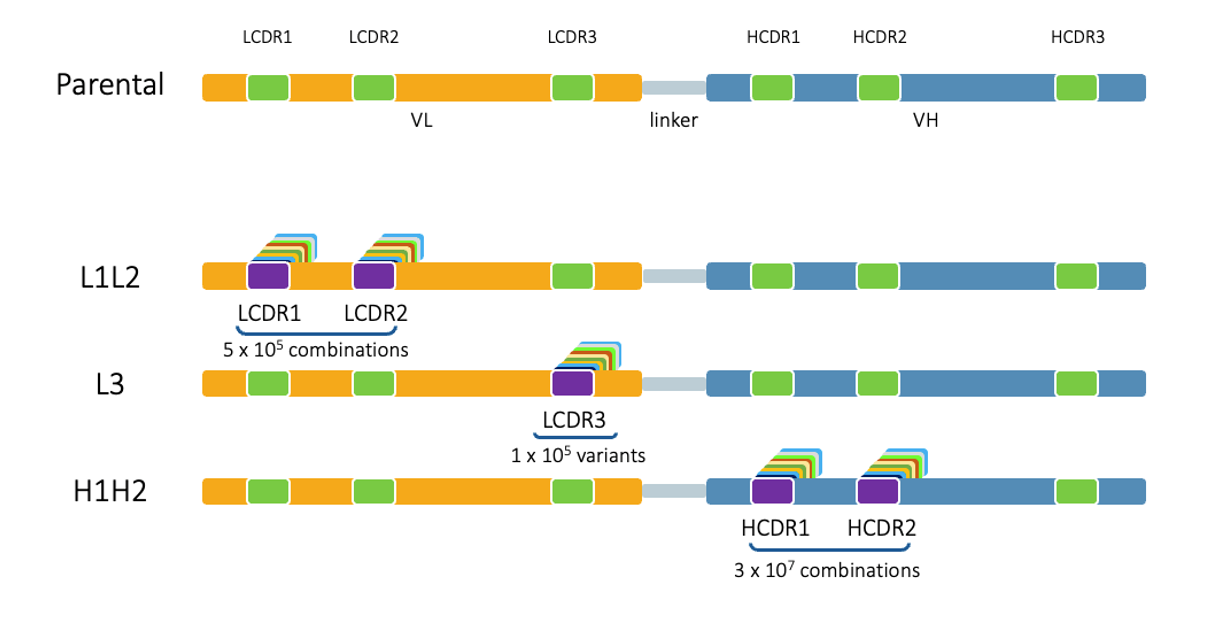
First, we create phase 1 libraries based on the antibody to be matured in which HCDR1/2, LCDR1/2, LCDR3, and HCDR3 if needed, are replaced with scaffold-matched CDRs from our Gen 3 platform.
Next, the outputs of phase one libraries are combined to create combination libraries, which undergo high stringency selection for desired properties.
Finally, selection outputs are evaluated by next-gen sequencing to remove sequence liabilities from the CDRs. The resulting affinity maturation ranges from 10-200 fold over the starting antibody affinity, while also improving developability.
Exploring the Gen 3 CDR Space
Sequential Approach: Phase 1 - Individual Sub-Libraries
Sequential Approach: Phase 2 - Combo Library
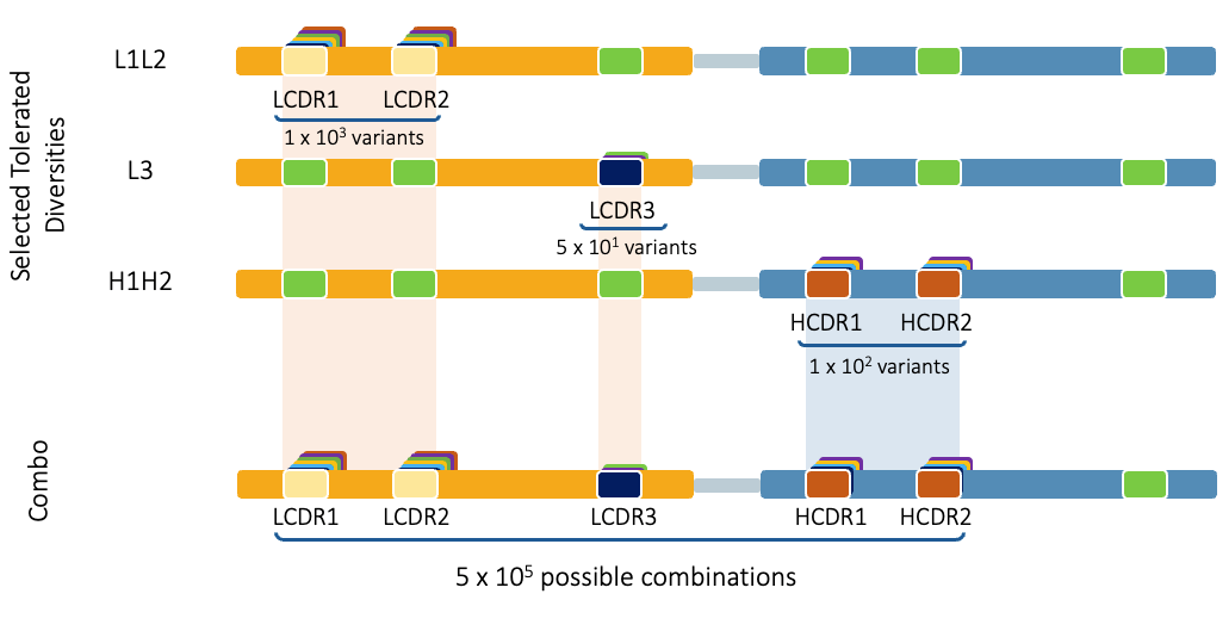

Generally, the more CDRs targeted for improvement, the greater the affinity improvement obtained.
Advantages of using Specifica’s Generation 3 platform for affinity maturation and developability optimization
Natural Diversity
Natural CDRs from Specifica’s in-house database are used in antibody improvement protocols, each matched to the framework of your antibody, improving the quality of your natural diversity.
Deliverables
For affinity maturation services, Specifica guarantees to improve the affinity of your antibodies by at least 10-fold, with preserved or improved developability properties.
All-human sequences
The use of only natural human CDRs matched to the germline of your antibody maintains or increases humanness.
Straightforward terms
Specifica’s payment terms add no royalties or milestone fees, with full payment only occurring upon the successful completion of a project.
No sequence liabilities
By eliminating all CDRs containing sequence liabilities from the diversity, Specifica simultaneously improves both developability and affinity.
Collaborate and customize
In addition to the in-house approach, Specifica also offers custom solutions based on customer input or structural knowledge. The parental HCDR3 sequence is typically retained but can also be mutated by scanning mutagenesis.
Two-stage procedure
This ensures the exploration of a diversity of up to 1018 CDR combinations while preserving epitope recognition.

